

ORAL BIOFILMS: ARCHITECTS OF DISEASE, HEAD TO TOE
In the oral microbiota, there are greater than 770 different microbial species; however, a select proportion will up-regulate from a planktonic phenotype to a biofilm phenotype , attached to a biotic surface or non biotic surfaces based on their stress response genes and ‘Quorum sensing’. Eight key environmental/cultural characteristics magnify the selection of multi-species biofilm inhabitants during a 4 -stage life cycle (I-IV), enhance by metastasis to distal sights: gingivitis (micro-aerophilic microbes), periodontitis and endodontics (anaerobic microbes).
The unifying concept is the Ecological or Plaque Hypothesis , (PD Marsh, Wales)which recognizes that ecological pressure (including antibiotics) is necessary for low number “Professional Pathogens” to outcompete resident flora , “Beneficial Microbes “ and achieve numerical dominance (ratio) associated with a biofilm phenotype . In our 2 working BIOFILM models (VAP and Wounds), this was identified as ETT, a dynamic biofilm engine, and wound bed as a static biofilm reactor, respectively.
Architecturally, biofilms promote the theme of “Structure equals Function”, incorporating structural characteristics that redefine a biofilm, acting as a hydrated polymer, focusing on chemical features including Rheology (Movement/Stationary) and (Viscoelastic) dispersal of energy. Biofilms include cross bridging for strength, nano-wires for communication, and HGT (Horizontal Gene Transfer), promoting antibiotic resistance. Fungi, mostly Candida albicans, act as ‘universal co-aggregates’, promoting linking thru the outer most protective structures, EPS (Extra Polymeric Substance), highlighting prokaryotic and eukaryotic functionality.
Biofilms can also be described as a type of primitive developmental biology, in which spatial organization of cells within the matrix optimizes nutritional resources. In this state it acts as an immobilized enzyme system, where stead state can be radically altered by applying physical forces, such as sheer.
Clinically, biofilms are markedly resistant to most antibiotic protocols designed to treat single cell associated infections, Planktonic phenotype. Using biofilm designed tools like the CBD, an MBEC, Minimal Biofilm Elimination Concentration, can be established for antibiotic resistance, measuring a 100 fold increase to planktonic isolate treatment using the MIC, Minimal Inhibitory Concentration for single cells. Multispecies are inherently resistant to antibiotic RX.
Hence, Therapeutic Modalities (sonication, silver ions/tobromycin, CMTs-chemically modified tetracyclines -and immune modulation) have refocused on multiple interventions, recognizing the properties of biofilms are similar to hydrated organic polymers, perhaps acting as tumors (multispecies communities) and the use of probiotics, Restorative Microbiology. We have combined these 2 pathways, recently, linking a “Disruptive:Reconstruction” concept of biofilm intervention, followed by probiotic reconstruction, recognizing the Plaque Hypothesis and Beneficial Microbes. (See Key Points, B. Biofilms and Oral health.)
BIOTUMORS or BIOFILMS: BENIGN TUMORS OR BIOFILMS AND TUMORS: A METASTATIC CONNECTION
Our passion for biofilm research also catalyzed ‘critical thinking’, thinking out-side the box. It became apparent to us that both human tumors, Eucaryotes, and biofilms, Procaryotes, were multi-system cellular organizations which shared at least 12 characteristics. It was apparent neoplasia’s, human, and biofilms, microbial, addressed multi-cellularity where metastasis was a common means of distribution, Stage II to Stage III. Together with Dr S. Shabahang, we then developed 2 hypotheses highlighting this link:
HYPOTHESIS I. Biofilms are prokaryotic Tumors (BIOTUMOR) where communities of linked microbes masquerading as a biofilm are really Procariotic tumors.
HYPOTHESIS II. Tumors are Eucaryotic Bioflms , where communities of similar linked cells masquerading as a tumor as really Eucaryotic Biofilms.
We developed a unique testing strategy, recognizing these similarities, linking Prokaryotic and Eucaryotic Therapy, predicting cancer therapy on biofilm response, comparing multiple tumor therapies. The evolution followed designing a concept, proof of concept with ultimately the creation of a BioTumor classification dependent upon biofilm response of 12 cancer drugs, emphasizing synergy.
Our evolution could be traced by abstract and poster presentations at international meetings:

The Center of Excellence in Biofilm Research (CEBR) is an innovative research environment. Working in close collaboration with clinicians and Carnegie Mellon faculty, the CEBR fosters the development of surgeon-scientists, physician-scientists, and biomedical researchers by actively working to develop partnerships among teams of individuals with diverse scientific, clinical, computational, and engineering backgrounds.
CEBR's goal is to promote human health by providing a deeper understanding of the role of microbial biofilms in the disease process. Currently researchers are conducting pioneering studies in the specialties of orthopaedics, cardiology, oncology, obstetrics, and urology. The goal of these collaborations is to better understand the role bacteria, particularly biofilms, play in complex clinical situations.
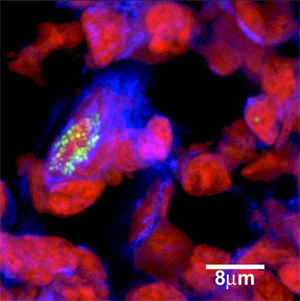
Bacteria in biofilms tend to be more difficult to culture and more resistant to control strategies (antibiotics and biocides) and host defenses than when grown planktonically in the laboratory. Biofilms are communities of micro-organisms encased within extracellular polymeric substances (EPS) living on surfaces. Their resilience has been related to physiology and protection by the EPS matrix that they produce. Biofilms are dynamic and the composite microbial communities change over time, moving, shedding, and re-growing adherent colonies. These phenomena may explain seemingly conflicting features of the disease when signs and symptoms are otherwise consistent with infection.
CEBR's Critical Areas of Interest:
In order to study biofilms, state of the art molecular technologies are necessary. The CEBR employs a suite of advanced molecular technologies to study biofilm populations. The Pathogen Pipeline not only identifies the microbial composition of a sample but provides visual proof of the pathogen's existence. The combination of the three technologies: PCR-ESI-TOF-MS, microbial sequencing, and FISH-confocal microscopy do not exist in combination at any other location. In addition the Center offers genomics, transcriptomics, proteomics, and bioinformatics to provide additional insights about the role of biofilms in the disease process and help us investigate our critical areas of interest.
Step One: Microbial Detection
Using cutting-edge DNA-based technology, PCR-ESI-TOF-MS, AHN CEBR scientists can identify specific strains of bacteria and fungi without the need for culture. The system combines three technologies that provide researchers with broad-based microbial diagnostics. First, this technology employs multiple PCR-based DNA amplifications targeted to highly conserved genes throughout the bacterial and fungal domains. Next, it employs a novel mass spectroscopic analysis termed ESI-TOF-MS that provides exact DNA base compositions for each of the PCR products. Finally, it utilizes a sophisticated mass look-up table and a computational triangulation approach to arrive at a strain-level diagnosis based on the presence/absence and weights of all of the amplified DNA targets.
In addition, microbial detection can be accomplished with the use of 16S ribosomal RNA (rRNA) sequencing available through CEBR. 16S ribosomal RNA (rRNA) sequencing provides another avenue to identify and compare bacteria present within a given sample. Sample barcoding allows for the simultaneous sequencing of multiple samples in a single run. Although more expensive than the PCR-ESI-TOF-MS technology, 16S rRNA gene sequencing can be used independently or as a confirmatory technology. Data from 16S studies can be used to improve the accuracy of bacterial classification.
Step Two: Microbial Visualization
The imaging of biofilms is fundamental to biofilm research. High-resolution confocal images provided the first evidence for the structural heterogeneity of biofilm architecture. This evidence was used to establish models of biofilm growth and ultrastructure. The use of confocal scanning laser microscopy and florescent in situ hybridization (FISH) on ex vivo clinical samples allows researchers the ability to detect and localize the presence or absence of specific nucleic acid sequences. Fluorescent probes bind to complementary sequences which can then be visualized using a confocal microscope. The technique is powerful not only in its ability to confirm the IBIS results but to visualize the location of the bacteria within a sample.
Step Three: Microbial Exploration
There are many other tools available to investigate biofilms. Here is a brief list of some of the other services we can provide.
1)Nickel JC, Stephens A, Landis JR, Chen J, Mullins C, van Bokhoven A, Lucia MS, Melton-Kreft R, Ehrlich GD; The MAPP Research Network. (2015) Search for Microorganisms in Men with Urologic Chronic Pelvic Pain Syndrome: A Culture-Independent Analysis in the MAPP Research Network. J Urol. Jul;194(1):127-35.
2)Kathju S, Nistico L, Melton-Kreft R, Lasko LA, Stoodley P.Direct demonstration of bacterial biofilms on prosthetic mesh after ventral herniorrhaphy. Surg Infect (Larchmt). 2015 Feb;16(1):45-53.
3)Palmer MP, Altman DT, Altman GT, Sewecke JJ, Ehrlich GD, Hu FZ, Nistico L, Melton-Kreft R, Gause TM 3rd, Costerton JW. Can we trust intraoperative culture results in nonunions? J Orthop Trauma. 2014 Jul;28(7):384-90.
4)Jacovides CL, Kreft R, Adeli B, Hozack B, Ehrlich GD,Parvizi J. Successful identification of pathogens by polymerase chain reaction (PCR)-based electron spray ionization time-of-flight mass spectrometry (ESI-TOF-MS) in culture-negative periprosthetic joint infection. J Bone Joint Surg Am. 2012 Dec 19;94(24):2247-54.
5)Yun HC, Kreft RE, Castillo MA, Ehrlich GD, Guymon CH,Crouch HK, Chung KK, Wenke JC, Hsu JR, Spirk TL, Costerton JW, Mende K, Murray CK. Comparison of PCR/electron spray ionization-time-of-flight-mass spectrometry versus traditional clinical microbiology for active surveillance of organisms contaminating high-use surfaces in a burn intensive care unit, an orthopedic ward and healthcare workers. BMC Infect Dis. 2012 Oct 10;12:252.
6)Tuttle MS, Mostow E, Mukherjee P, Hu FZ, Melton-KreftR, Ehrlich GD, Dowd SE, Ghannoum MA. Characterization of bacterial communities in venous insufficiency wounds by use of conventional culture and molecular diagnostic methods. J Clin Microbiol. 2011 Nov;49(11):3812-9.
7)Kreft, R., Costerton, J. W., & Ehrlich, G. D. . PCR ischanging clinical diagnostics. Microbe 2010; 8(1):15-20.
8)Hall-Stoodley L, Hu FZ, Gieseke A, Nistico L, Nguyen D,Hayes J, Forbes M, Greenberg DP, Dice B, Burrows A, Wackym PA, Stoodley P, Post JC, Ehrlich GD, Kerschner JE. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA. 2006 Jul 12;296(2):202-11.